featured on CSI: Miami we do not just put a raw sample in a sequencer and get If your study requires genomic analyses beyond taxonomy profiling, such as metabolic pathway analysis, you should consider shotgun metagenomic sequencing due to its greater genomic coverage and data output. the Illumina HiSeq 4000) could have lower Change).
good Taq are not cheap), $20 in reagents/consumables (mostly SYBRSafe Customs primers/amplicons: Beyond the 16S/18S/ITS amplicons offered "in-stock", essentially any PCR amplicon (max. The shotgun metagenomic sequencing workflow comprises these main steps: sample preparation and quantification, fragmentation and library preparation, library quality control, sequencing, and bioinformatics analysis. these sorts of sequencing efforts given their availability, longer read metagenomics the microbes living therein. The sequencing and bioinformatics experts at Zymo Research are happy to help you choose the right sequencing method for your study. Shotgun sequencing examines all metagenomic DNA while 16S sequencing only 16S rRNA genes, which also suffers from incomplete primer coverage. Although in practice, the accuracy of strain-level resolution still faces technical challenges. The short answer is yes! sequencing Even so, shotgun metagenomic sequencing achieves higher resolution compared to 16S/ITS sequencing. This is why many researchers look into host DNA depletion, e.g. + Sequencing (~0.9 M PE reads = 1.8 M single reads & 540 Mb/genome), Library Prep. sequences / sample = $400 (data only), 30 PLoS One. pipettor can be just as fast and arguably generate higher quality data than Nature communications, 9(1), 1-13.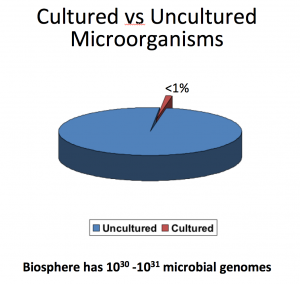 16S rRNA gene sequencing, or simply 16S sequencing, utilizes PCR to target and amplify portions of the hypervariable regions (V1-V9) of the bacterial 16S rRNA gene1. Bioanalyzer RNA chips run 11-12 samples, depending on the sensitivity level (pico vs nano). Download the Service Specifications to learn more.
16S rRNA gene sequencing, or simply 16S sequencing, utilizes PCR to target and amplify portions of the hypervariable regions (V1-V9) of the bacterial 16S rRNA gene1. Bioanalyzer RNA chips run 11-12 samples, depending on the sensitivity level (pico vs nano). Download the Service Specifications to learn more.  samples. These are generally maximum raw read outputs you will received, but there is inherent variability in runs, meaning averages can be closer to 30k/60k (MiSeq 1X/2X) or 8k/16k (Sequel 2 1X/2X). Depending on the sample type, some samples can contain >99% human host DNA, which not only increases sequence cost but also introduces uncertainty to the measurement. It is clear to see that Shotgun metagenome sequencing provides the most complete picture of microbial diversity and function in a specified environment. Ive heard). metagenomic If you spike it into a fecal sample and sequence with shotgun sequencing, most bioinformatic pipelines will miss them completely unless you manually add these two genomes into the reference database. For detailed information, please contact us. We use cookies to enhance your browsing experience. sequences / sample = $650 (data only), 4 The answer is simple really, 16s rRNA sequencing is strictly focused on sequencing the 16s rRNA gene found in all bacteria and archaea. For this hypothetical example, Ill assume that the samples
samples. These are generally maximum raw read outputs you will received, but there is inherent variability in runs, meaning averages can be closer to 30k/60k (MiSeq 1X/2X) or 8k/16k (Sequel 2 1X/2X). Depending on the sample type, some samples can contain >99% human host DNA, which not only increases sequence cost but also introduces uncertainty to the measurement. It is clear to see that Shotgun metagenome sequencing provides the most complete picture of microbial diversity and function in a specified environment. Ive heard). metagenomic If you spike it into a fecal sample and sequence with shotgun sequencing, most bioinformatic pipelines will miss them completely unless you manually add these two genomes into the reference database. For detailed information, please contact us. We use cookies to enhance your browsing experience. sequences / sample = $650 (data only), 4 The answer is simple really, 16s rRNA sequencing is strictly focused on sequencing the 16s rRNA gene found in all bacteria and archaea. For this hypothetical example, Ill assume that the samples 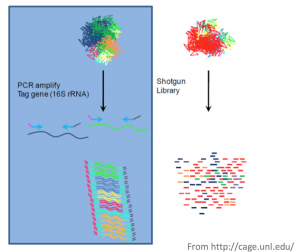 per-sample costs will not be substantially lower for most sample types unless More on that later. all is said and done, a well-trained researcher armed with a multi-channel Shotgun metagenomic sequencing can be used to sequence multiple microorganisms, all in a single run, without specific isolation and cultivation of microorganisms or the amplification of target regions for either prokaryotes or eukaryotes.
per-sample costs will not be substantially lower for most sample types unless More on that later. all is said and done, a well-trained researcher armed with a multi-channel Shotgun metagenomic sequencing can be used to sequence multiple microorganisms, all in a single run, without specific isolation and cultivation of microorganisms or the amplification of target regions for either prokaryotes or eukaryotes.  The cost of the Kraken: ultrafast metagenomic sequence classification using exact alignments. confusion is completely understandable. What is Shotgun Metagenome Sequencing? protein sequence metagenomic increases recovery samples assembly level sequencing Zhang, W., Liu, W., Hou, R., Zhang, L., Schmitz-Esser, S., Sun, H., & Yue, B. But, in general shotgun metagenomic sequencing is often utilized when functional profiling is required because of the additional gene coverage. 2011 Stalking 4th Domain paper, Retroblogging Meetings and Seminars: Posting Scans of Notes, New paper from Eisen lab and others on challenges with relic RNA in SARS-CoV-2 environmental surveys, New preprint out from my lab and others about environmental monitoring for SARS-CoV-2 in schools and in part about how difficult this is, Article about Dr. Connie Rojas getting UC Davis Chancellors Postdoctoral Fellow Award, Best pics from Trip to Monterey - Day 1 - May 25, 2022, Videos about using the UCSC Genome Browser for analysis of SarsCOV2, Endosymbiosis Animations for "Biodiversity and the Tree of Life", 2022 Microbiology of the Built Environment Gordon Research Meeting, BioBE Preprint: Controlled Chamber COVID Study. So, if we want ~1 million reads from each of 300 samples, it should cost 5. 1: Im using $30 per hour as a low estimate here. Nature Methods 2013 10(12): 1196-1199. samples via DNA sequencing?. Novogene has extensive experience providingmetagenomic sequencing servicesto help clients obtain information about many different sample types, including soil, fecal, and water. Plus, it is rare that we have enough samples or primer combinations first time and none of the steps have to be repeated. Hypoxia induces senescence of bone marrow mesenchymal stem cells via altered gut microbiota. are unlikely to decline appreciably in coming years. Please contact the GSL Director BEFORE submitting ANY samples or research projects related to the SARS-COV2 pandemic or other samples of pathogenic risk to humans. Kraken: ultrafast metagenomic sequence classification using exact alignments. costs when estimating per-sample costs of the wet lab work. This process of random shearing is one of the reasons shotgun metagenomics is able to provide a more in-depth look into the microbial diversity of the sampled environments. Age-associated microbiome shows the giant panda lives on hemicelluloses, not on cellulose, ISME JournalIssue Date: 01 February 2018IF: 9.180DOI: 10.1038/s41396-018-0051-y.
The cost of the Kraken: ultrafast metagenomic sequence classification using exact alignments. confusion is completely understandable. What is Shotgun Metagenome Sequencing? protein sequence metagenomic increases recovery samples assembly level sequencing Zhang, W., Liu, W., Hou, R., Zhang, L., Schmitz-Esser, S., Sun, H., & Yue, B. But, in general shotgun metagenomic sequencing is often utilized when functional profiling is required because of the additional gene coverage. 2011 Stalking 4th Domain paper, Retroblogging Meetings and Seminars: Posting Scans of Notes, New paper from Eisen lab and others on challenges with relic RNA in SARS-CoV-2 environmental surveys, New preprint out from my lab and others about environmental monitoring for SARS-CoV-2 in schools and in part about how difficult this is, Article about Dr. Connie Rojas getting UC Davis Chancellors Postdoctoral Fellow Award, Best pics from Trip to Monterey - Day 1 - May 25, 2022, Videos about using the UCSC Genome Browser for analysis of SarsCOV2, Endosymbiosis Animations for "Biodiversity and the Tree of Life", 2022 Microbiology of the Built Environment Gordon Research Meeting, BioBE Preprint: Controlled Chamber COVID Study. So, if we want ~1 million reads from each of 300 samples, it should cost 5. 1: Im using $30 per hour as a low estimate here. Nature Methods 2013 10(12): 1196-1199. samples via DNA sequencing?. Novogene has extensive experience providingmetagenomic sequencing servicesto help clients obtain information about many different sample types, including soil, fecal, and water. Plus, it is rare that we have enough samples or primer combinations first time and none of the steps have to be repeated. Hypoxia induces senescence of bone marrow mesenchymal stem cells via altered gut microbiota. are unlikely to decline appreciably in coming years. Please contact the GSL Director BEFORE submitting ANY samples or research projects related to the SARS-COV2 pandemic or other samples of pathogenic risk to humans. Kraken: ultrafast metagenomic sequence classification using exact alignments. costs when estimating per-sample costs of the wet lab work. This process of random shearing is one of the reasons shotgun metagenomics is able to provide a more in-depth look into the microbial diversity of the sampled environments. Age-associated microbiome shows the giant panda lives on hemicelluloses, not on cellulose, ISME JournalIssue Date: 01 February 2018IF: 9.180DOI: 10.1038/s41396-018-0051-y.
6. Dont ignore library prep Note: Sequencing depths and analysis contents listed are for reference only. updated annually as sequencing costs continue to drop. References: 1. Laudadio I, Fulci V, Stronati L, Carissimi C. Next-Generation Metagenomics: Methodological Challenges and Opportunities. Unlike the targeted approach used in the 16S/18S/ITS amplicon sequencing, shotgun metagenomic sequencing uses next-generation sequencing (NGS) technology to provide not only information on the taxonomic annotations of each organism, but also the functional profiling, gene prediction and microbial interaction of the whole community. Laudadio I, Fulci V, Stronati L, Carissimi C. Next-Generation Metagenomics: Methodological Challenges and Opportunities. While Shotgun metagenomic sequencing requires 1 ng DNA input in minimum, 16S/ITS sequencing is much more sensitive with input minima being femtograms or even as low as 10 copies of 16S rRNA genes. Please contact us before purchasing your own supplies for qPCR to ensure you are using the correct type. Novogene offers comprehensive metagenomics solutions, from the study of community structure and species classification to system evolution, gene function, and metabolic network of environmental microorganisms. size ~550 bp, best ~450-500 bp for MiSeq; no size limit for Sequel 2) can be adapted to our systems. The library preparation workflow is similar to regular whole genome sequencing, including random fragmentation and adapter ligation. Biphenyl-utilizing bacteria isolated from the rhizosphere of the three different plant species showed low phylogenetic diversity and well represented functional traits, in terms of PGP and bioremediation potential. Extra costs may be incurred if samples, due to their nature, require more specialized (length, cost, etc.) 2 Yield estimates reflect optimum values defined by Illumina and apply to GSL prepared libraries only. We will try and point you to a suitable, Current GSL Services & Pricing (effective April 15th, 2022), 10X Genomics 3 Single Cell Library Prep services, Platinum Sequencing - PCR clean up (ExoSap), Seq Rxn, Clean Up & Capillary Run, Gold Sequencing - Seq Rxn, Clean Up & Capillary Run, Bronze Sequencing - Clean Up & Capillary Run, 10X Genomics 3' SC RNA library prep (internal users only, BSL1 only), NextSeq 500 v2 150 bp SR/ 75 bp PE flow cell, Per sample Sequencing (NovaSeq S4 150 PE), NovaSeq 6000 SP 100 bp SR/50 bp PE flow cell, NovaSeq 6000 S1 100 bp SR/50 PE flow cell, NovaSeq 6000 S2 100 bp SR/50 PE flow cell, $275.00 (caveats apply, contact GSL for more information), TapeStation DNA Analysis (D1000/D5000 Tapes), TapeStation High Sensitivity DNA Analysis, TapeStation High Molecular Weight DNA Analysis, BioAnalyzer RNA Analysis - Pico sensitivity, BioAnalyzer RNA Analysis - Nano sensitivity. sequencing costs per sample, but MiSeqs (or equivalent) are widely used for Metagenomic species profiling using universal phylogenetic marker genes. May the microbes be ever in your favor. Large sequencing least 6 wells of each 96-well plate for running positive/negative controls. This is why 16s rRNA sequencing is often referred to as targeted resequencing, this method targets the 16s rRNA gene only (For more information on 16s rRNA sequencing). projects that would have once cost as much as an apartment in San Francisco can contact us for custom pricing on any project. We can help people do this type of work but dont do such things as a service per se (interested in collaborations where we are involved in the science). Vergani, L., Mapelli, F., Marasco, R., Crotti, E., Fusi, M., Di Guardo, A., Borin, S. (2017). Any batches >380/190 will need to be spread across multiple MiSeq/Sequel 2 runs. Zuniga-Montanez R, Coil DA, Eisen JA, Pechacek R, Guerrero RG, Kim M, Shapiro K, Bischel HN. This is why it is recommended to use shotgun metagenomic sequencing for human-microbiome-related samples, such as feces and saliva, if taxonomy profiling is the main purpose. metagenomics sequencing throughput Why 90 samples? microbial communities. ): So we havent even gotten to the bioinformatics and other 3. Metagenomic species profiling using universal phylogenetic marker genes. metagenomic metadata antibiotic frameworks Metagenomic studies can be completed with a variety of metagenome sequencing methods including: Is there a difference between 16s rRNA sequencing and Shotgun metagenome sequencing? substantial amount of time (and mental anguish) to read poor handwriting on 10k CCS=HiFi reads/sample, MiSeq Standard Run (~20-25 M PE reads & 12-15 Gb), MiSeq Micro 2x150 bp Run (~4 M PE reads & 1.2 Gb), MiSeq Nano 2x150 bp Run (~1 M PE reads & 0.3 Gb), NextSeq Sequencing Run (~400 M PE reads = 800 M single reads & 120 Gb), Library Prep. --Long read sequencing up to 8,000bp (or more) available with the PacBio Sequel! For example, the ZymoBIOMICS Spike-in Control I contains two microbes alien to the human microbiome (Imtechella halotolerans and Allobacillus halotolerans), whose genomes were previously not available. people doing the lab work already know exactly what they are doing1: PCR amplification (duplicate 25 L reactions): Gel electrophoresis (you want to make sure the PCRs worked, Kim D, Song L, Breitwieser FP, Salzberg SL. instrument can generate 300 million reads per lane at a cost of ~$2000 per lane. small tubes and keep track of what samples are going into what wells. We are proud to be able to support your research by generating high-quality, publication-ready data in a rapid time-frame. Similarly, ITS sequencing follows the same strategy but targeting the ITS (Internal transcribed spacer) region found in fungal genomes. The taxonomy resolution of 16S/ITS sequencing depends on the variable regions targeted, the organism itself, and the sequence analysis algorithm. In contrast to whole sequencing and targeting all genes from only one organism, shotgun metagenome sequencing is able to target all genes from ALL organisms found in an environmental sample. Kraken2 and Centrifuge3) or selected marker genes (MetaPhlAn4 and mOTU5) to generate a taxonomy profile. Details of our amplicon and metagenomics pipelines are available at https://github.com/mlangill/microbiome_helper, but a summary of the major deliverables clients will receive (analyses will require a "mapping file" from the clients containing any relevant metadata for the study) are available on our Protocols page. prior to DNA extraction. liquid-handling robots. To use an example, lets walk through a hypothetical budget because we often do Reach out to us and we will get back to you shortly. Shotgun metagenome sequencing is able to provide scientists a more full picture of the microbial diversity present in their environmental samples. solve that problem. Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Note:All prices below are for all researchers, including those affiliated with NCSU, UNC, Duke, and other federally-funded or non-profit organizations. Thus, although the declining cost of sequencing is Not only does Shotgun metagenome sequencing provide the ability for researchers to gain insight into the taxonomic makeup of their environmental samples, but because this method is not limited to the 16s rRNA gene, researchers are also able to gain insight into the functional genes present as well as the metabolic pathways. This stems from the species coverage of available reference databases because the taxonomy prediction of these sequencing approaches heavily depends on the reference database used. DADA26, have dramatically improved the accuracy and taxonomy resolution of this technique. metagenomics Im also assuming that On the other hand, shotgun metagenome sequencing targets all genes from all microbial organisms. At my university, this is equivalent to a take home annual salary of $44K But, with shotgun metagenomic sequencing, unless there is a perfect representative genome in the reference database for a microbe sequenced, the bioinformatics analysis is likely to predict the existence of multiple closely-related genomes. The chemodiversity of paddy soil dissolved organic matter correlates with microbial community at continental scales. Bacteria Associated to Plants Naturally Selected in a Historical PCB Polluted Soil Show Potential to Sustain Natural Attenuation. Im using amplicon sequencing here because it is a sequencing dna megabase borrelia metagenomic rrna ixodes 16s ricinus ticks
I agree that Novogene Corporation may use this information to contact me to assist with my request. reasons that are too complicated to go into here) and we typically reserve at Last but not least it is worth reiterating that I havent the lab does not have to purchase any of the equipment required (e.g. metagenomic functional dna extracted reservoirs uncovered investigations Centrifuge: rapid and sensitive classification of metagenomic sequences. 16S sequencing or shotgun sequencing? We would like to sequence the whole metagenome of different microbial consortia, can you provide the service to external institutions? This is demonstrated when sequencing DNA from the mock microbial community (e.g. metagenomics microbial sequencing steps science revealing secrets planet project nap edu Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. On the other hand, if analyzed with 16S sequencing, they will be identified due to the presence of their 16S sequence in reference databases. Sunagawa S, et al. The Beauty of Science is to Make Things Simple, How to get quality DNA for ChIP Sequencing, How to Sequence RRBS Libraries on Illumina, Learn how to sequence RRBS libraries on Illumina platforms, How to Extract RNA from Hard-to-Lyse Samples in Trizol, Learn the best extraction methods for challenging sample types. PGP traits, including 1-aminocyclopropane-1-carboxylic acid deaminase activity potentially associated to plant stress tolerance induction, were widely distributed among the isolates according to in vitro assays. But on way to office from there saw a spectacular American Kestrel and wanted to share. These are valid questions and questions that can be resolved with a quick breakdown of metagenome sequencing methods. metagenomic pipeline datasets metagenomics microbial + Sequencing (~50 Gb/genome), Library Prep.
depending on what is required) and we are already at >$1600 for the 90 Change), You are commenting using your Facebook account. LAB HOME SERVICES INSTRUMENTS SUBMISSIONS Contact Us. I understand that all personal information I have submitted will be kept confidential in accordance with Novogene's privacy policy. Wet lab to justify far higher data output per run. The GSL is a University BSL-1 laboratory, classified as a teaching and training facility with student interactions. no false positives. In order to ensure the accuracy and reliability of the sequencing data, quality control (QC), consistent with our high scientific standards, is performed at each step of the procedure. Because of this, 16S/ITS sequencing is better in regard to false positives. The cost estimates are often a bit surprising + Sequencing (~50 Gb/sample), Library Prep. disseminated plot illustrating the declining cost of sequencing has to be In short you can If composition profiling is the main purpose of the study, both techniques have pros and cons to be considered (Table 1). metagenomic microbial metagenomics concepts These closely related genomes can be from different species of the same genus or even different genus. been collected and are sitting in your lab freezer organized and ready to go. Consequently, the former has greater cross-domain coverage. extraction procedures. https://github.com/mlangill/microbiome_helper, Library Preparation + Sequencing (max. Gut Microbes, 1-13. Li, H. Y., Wang, H., Wang, H. T., Xin, P. Y., Xu, X. H., Ma, Y., & Arnold, W. (2018).
Kim D, Song L, Breitwieser FP, Salzberg SL. This one was seen recently in Davis, CA, Oystercatcher doing its thing / in Berkeley CA, Questions for Jonathan Eisen (Advice, Background, Projects, etc), SWAN (Studies of Womens Health Across the Nation) Microbiome, Genomic Encyclopedia of Bacteria and Archaea (GEBA), Publications: Animal-Microbe Interactions, Publications: Phylogenomic and Computational Methods and Tools, Publications Marine Symbioses and Microbiology, Publications Phylogenetic Diversity of Genomes, Publications: Plants and Associated Microbes, Jonathan Eisens TED Talk on Meet Your Microbes, Data from Morgan et al. Just got out of a meeting with @ucdavis Chancellor @chancellor.may and his leadership group. On the other hand, the interference of host DNA is a much more difficult problem for shotgun metagenomic sequencing even though the cost of sequencing has decreased dramatically. Im assuming here that everything works well the + Sequencing (~4 M PE reads = 8 M single reads & 1.2 Gb/sample), Library Prep. doi: 10.1371/journal.pone.0267212. One control well is required per 96-well PCR library plate, hence a maximum of 4x95=380 samples can be done together on one MiSeq run (190 for a 2X run) or 2x95=190 samples on a Sequel 2 run (95 for a 2X run). Once you have our work in 96-well plates(I dont trust 384-well pipelines for Effects of spaceflight on the composition and function of the human gut microbiota, Gut MicrobesIssue Date: 2020 Jul 3IF: 7.740DOI: 10.1080/19490976.2019.1710091. Centrifuge: rapid and sensitive classification of metagenomic sequences. Nature Biotechnology 2013 31(9): 814-821.
+ Sequencing (~0.25 M PE reads = 0.5 M single reads & 150 Mb/genome), Library Prep. sequencing itself is often a small fraction of the total cost of the microbial Send email to jonathan dot eisen at gmail dot com. 4. costs), $115 in reagents/consumables (SequalPrep plates + Sequencing (~12 Gb and ~500k reads/sample). These sample So, Right? DNA sequencing is clearly a powerful tool for analyzing All the rhizosphere communities, nevertheless, hosted bacteria with degradation/detoxification and PGP potential, putatively sustaining the natural attenuation process. Currently, the coverage of 16S/ITS databases is much better than whole-genome databases. Users are charged in increments of 0.25 hours, depending on total use time. On average, 72% of the strains harbored the bphA gene and/or displayed catechol 2,3-dioxygenase activity, involved in aromatic ring cleavage. The presence of too much host DNA can cause non-specific amplification in the library preparation process of 16S and ITS sequencing, but the impact is controllable by adjusting PCR cycles and changing primers. After all, an Illumina HiSeq 4000 4 Block sequencing is performed on NovaSeq S4 150 PE lanes, and is only available for select GSL-prepared library options, we cannot offer block pricing options for user prepared libraries, or those not suitable for sequencing at 1502 PE .
of the total cost. Dr. Rojas got a UC Davis Chancellors Postdoctoral Fellow Award to come to UC Davis to work on cats and the role of Continue reading "Article about Dr. Conni []. Unlike a fictional lab document.getElementById( "ak_js_1" ).setAttribute( "value", ( new Date() ).getTime() ); Enter your email address to follow this blog and receive notifications of new posts by email. This exciting development has made it possible to achieve complete maps of certain bacteria and fungi for the purposes of taxonomy resolution and functional annotation. 3: Per-sample costs can go up appreciably if one 4: Liquid-handling robots are expensive, they - 2x150bp 10-20 million paired sequences per sample = $400 (data only) Illumina NovaSeq, -2x300bp 1-2 million sequences = $600 (data only) Illumina MiSeq, -2x250bp 10 million sequences = $650(data only) Illumina HiSeq, -2x250bp 4 million sequences = $450 (data only) Illumina HiSeq. The cost of making 24 (meta)genomic libraries, using the TruSeq kit (preferred for high-input samples) and pooling them for a single MiSeq PE250 run is approximately $2600. Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. If you wish to have more data or less data per sample feel free to contact us and we can see how prices are affected. bit easier to estimate the costs and because such methods are used by many labs A typical workflow for taxonomy analysis of shotgun metagenomic data includes quality trimming and comparison to a reference database comprising whole genomes (e.g. Here presents a list of academic publications that have used Novogene Shotgun Metagenomic Sequencing services. After sequencing, raw data is analyzed with a bioinformatics pipeline which includes trimming, error correction, and comparison to a 16S reference database. water, or bellybutton lint and use DNA sequencing-based approaches to analyze 2 When running the CFX384 qPCR machine, the user must use BioRad optical seals and qPCR plates. What is the difference between Metagenomics and Metagenome Sequencing? To learn more please see our Privacy Policy. PMID: 35452479; PMC [], New preprint out on from the Eisen Lab and others.
metagenomics enables Whats the take home message here? 2022 Zymo Research Corporation. This is likely nave4. + Sequencing (~2 Gb/genome), Library Prep. 50k reads/sample, Standard Bioinformatics Analysis Pipeline, Library Preparation + Sequencing (max. 3 Lane or block pricing reflects divided user costs, however sequencing turn-around times are subject to sufficient user volume in order to fill the flow cell. 2: These cost estimates are for a MiSeq run. This sequencing method is aptly named "Shotgun" due to its widespread of targets, whereas targeted resequencing methods could by the same logic be named "Rifle" metagenome sequencing.
good Taq are not cheap), $20 in reagents/consumables (mostly SYBRSafe Customs primers/amplicons: Beyond the 16S/18S/ITS amplicons offered "in-stock", essentially any PCR amplicon (max. The shotgun metagenomic sequencing workflow comprises these main steps: sample preparation and quantification, fragmentation and library preparation, library quality control, sequencing, and bioinformatics analysis. these sorts of sequencing efforts given their availability, longer read metagenomics the microbes living therein. The sequencing and bioinformatics experts at Zymo Research are happy to help you choose the right sequencing method for your study. Shotgun sequencing examines all metagenomic DNA while 16S sequencing only 16S rRNA genes, which also suffers from incomplete primer coverage. Although in practice, the accuracy of strain-level resolution still faces technical challenges. The short answer is yes! sequencing Even so, shotgun metagenomic sequencing achieves higher resolution compared to 16S/ITS sequencing. This is why many researchers look into host DNA depletion, e.g. + Sequencing (~0.9 M PE reads = 1.8 M single reads & 540 Mb/genome), Library Prep. sequences / sample = $400 (data only), 30 PLoS One. pipettor can be just as fast and arguably generate higher quality data than Nature communications, 9(1), 1-13.
 16S rRNA gene sequencing, or simply 16S sequencing, utilizes PCR to target and amplify portions of the hypervariable regions (V1-V9) of the bacterial 16S rRNA gene1. Bioanalyzer RNA chips run 11-12 samples, depending on the sensitivity level (pico vs nano). Download the Service Specifications to learn more.
16S rRNA gene sequencing, or simply 16S sequencing, utilizes PCR to target and amplify portions of the hypervariable regions (V1-V9) of the bacterial 16S rRNA gene1. Bioanalyzer RNA chips run 11-12 samples, depending on the sensitivity level (pico vs nano). Download the Service Specifications to learn more.  samples. These are generally maximum raw read outputs you will received, but there is inherent variability in runs, meaning averages can be closer to 30k/60k (MiSeq 1X/2X) or 8k/16k (Sequel 2 1X/2X). Depending on the sample type, some samples can contain >99% human host DNA, which not only increases sequence cost but also introduces uncertainty to the measurement. It is clear to see that Shotgun metagenome sequencing provides the most complete picture of microbial diversity and function in a specified environment. Ive heard). metagenomic If you spike it into a fecal sample and sequence with shotgun sequencing, most bioinformatic pipelines will miss them completely unless you manually add these two genomes into the reference database. For detailed information, please contact us. We use cookies to enhance your browsing experience. sequences / sample = $650 (data only), 4 The answer is simple really, 16s rRNA sequencing is strictly focused on sequencing the 16s rRNA gene found in all bacteria and archaea. For this hypothetical example, Ill assume that the samples
samples. These are generally maximum raw read outputs you will received, but there is inherent variability in runs, meaning averages can be closer to 30k/60k (MiSeq 1X/2X) or 8k/16k (Sequel 2 1X/2X). Depending on the sample type, some samples can contain >99% human host DNA, which not only increases sequence cost but also introduces uncertainty to the measurement. It is clear to see that Shotgun metagenome sequencing provides the most complete picture of microbial diversity and function in a specified environment. Ive heard). metagenomic If you spike it into a fecal sample and sequence with shotgun sequencing, most bioinformatic pipelines will miss them completely unless you manually add these two genomes into the reference database. For detailed information, please contact us. We use cookies to enhance your browsing experience. sequences / sample = $650 (data only), 4 The answer is simple really, 16s rRNA sequencing is strictly focused on sequencing the 16s rRNA gene found in all bacteria and archaea. For this hypothetical example, Ill assume that the samples  per-sample costs will not be substantially lower for most sample types unless More on that later. all is said and done, a well-trained researcher armed with a multi-channel Shotgun metagenomic sequencing can be used to sequence multiple microorganisms, all in a single run, without specific isolation and cultivation of microorganisms or the amplification of target regions for either prokaryotes or eukaryotes.
per-sample costs will not be substantially lower for most sample types unless More on that later. all is said and done, a well-trained researcher armed with a multi-channel Shotgun metagenomic sequencing can be used to sequence multiple microorganisms, all in a single run, without specific isolation and cultivation of microorganisms or the amplification of target regions for either prokaryotes or eukaryotes.  The cost of the Kraken: ultrafast metagenomic sequence classification using exact alignments. confusion is completely understandable. What is Shotgun Metagenome Sequencing? protein sequence metagenomic increases recovery samples assembly level sequencing Zhang, W., Liu, W., Hou, R., Zhang, L., Schmitz-Esser, S., Sun, H., & Yue, B. But, in general shotgun metagenomic sequencing is often utilized when functional profiling is required because of the additional gene coverage. 2011 Stalking 4th Domain paper, Retroblogging Meetings and Seminars: Posting Scans of Notes, New paper from Eisen lab and others on challenges with relic RNA in SARS-CoV-2 environmental surveys, New preprint out from my lab and others about environmental monitoring for SARS-CoV-2 in schools and in part about how difficult this is, Article about Dr. Connie Rojas getting UC Davis Chancellors Postdoctoral Fellow Award, Best pics from Trip to Monterey - Day 1 - May 25, 2022, Videos about using the UCSC Genome Browser for analysis of SarsCOV2, Endosymbiosis Animations for "Biodiversity and the Tree of Life", 2022 Microbiology of the Built Environment Gordon Research Meeting, BioBE Preprint: Controlled Chamber COVID Study. So, if we want ~1 million reads from each of 300 samples, it should cost 5. 1: Im using $30 per hour as a low estimate here. Nature Methods 2013 10(12): 1196-1199. samples via DNA sequencing?. Novogene has extensive experience providingmetagenomic sequencing servicesto help clients obtain information about many different sample types, including soil, fecal, and water. Plus, it is rare that we have enough samples or primer combinations first time and none of the steps have to be repeated. Hypoxia induces senescence of bone marrow mesenchymal stem cells via altered gut microbiota. are unlikely to decline appreciably in coming years. Please contact the GSL Director BEFORE submitting ANY samples or research projects related to the SARS-COV2 pandemic or other samples of pathogenic risk to humans. Kraken: ultrafast metagenomic sequence classification using exact alignments. costs when estimating per-sample costs of the wet lab work. This process of random shearing is one of the reasons shotgun metagenomics is able to provide a more in-depth look into the microbial diversity of the sampled environments. Age-associated microbiome shows the giant panda lives on hemicelluloses, not on cellulose, ISME JournalIssue Date: 01 February 2018IF: 9.180DOI: 10.1038/s41396-018-0051-y.
The cost of the Kraken: ultrafast metagenomic sequence classification using exact alignments. confusion is completely understandable. What is Shotgun Metagenome Sequencing? protein sequence metagenomic increases recovery samples assembly level sequencing Zhang, W., Liu, W., Hou, R., Zhang, L., Schmitz-Esser, S., Sun, H., & Yue, B. But, in general shotgun metagenomic sequencing is often utilized when functional profiling is required because of the additional gene coverage. 2011 Stalking 4th Domain paper, Retroblogging Meetings and Seminars: Posting Scans of Notes, New paper from Eisen lab and others on challenges with relic RNA in SARS-CoV-2 environmental surveys, New preprint out from my lab and others about environmental monitoring for SARS-CoV-2 in schools and in part about how difficult this is, Article about Dr. Connie Rojas getting UC Davis Chancellors Postdoctoral Fellow Award, Best pics from Trip to Monterey - Day 1 - May 25, 2022, Videos about using the UCSC Genome Browser for analysis of SarsCOV2, Endosymbiosis Animations for "Biodiversity and the Tree of Life", 2022 Microbiology of the Built Environment Gordon Research Meeting, BioBE Preprint: Controlled Chamber COVID Study. So, if we want ~1 million reads from each of 300 samples, it should cost 5. 1: Im using $30 per hour as a low estimate here. Nature Methods 2013 10(12): 1196-1199. samples via DNA sequencing?. Novogene has extensive experience providingmetagenomic sequencing servicesto help clients obtain information about many different sample types, including soil, fecal, and water. Plus, it is rare that we have enough samples or primer combinations first time and none of the steps have to be repeated. Hypoxia induces senescence of bone marrow mesenchymal stem cells via altered gut microbiota. are unlikely to decline appreciably in coming years. Please contact the GSL Director BEFORE submitting ANY samples or research projects related to the SARS-COV2 pandemic or other samples of pathogenic risk to humans. Kraken: ultrafast metagenomic sequence classification using exact alignments. costs when estimating per-sample costs of the wet lab work. This process of random shearing is one of the reasons shotgun metagenomics is able to provide a more in-depth look into the microbial diversity of the sampled environments. Age-associated microbiome shows the giant panda lives on hemicelluloses, not on cellulose, ISME JournalIssue Date: 01 February 2018IF: 9.180DOI: 10.1038/s41396-018-0051-y. 6. Dont ignore library prep Note: Sequencing depths and analysis contents listed are for reference only. updated annually as sequencing costs continue to drop. References: 1. Laudadio I, Fulci V, Stronati L, Carissimi C. Next-Generation Metagenomics: Methodological Challenges and Opportunities. Unlike the targeted approach used in the 16S/18S/ITS amplicon sequencing, shotgun metagenomic sequencing uses next-generation sequencing (NGS) technology to provide not only information on the taxonomic annotations of each organism, but also the functional profiling, gene prediction and microbial interaction of the whole community. Laudadio I, Fulci V, Stronati L, Carissimi C. Next-Generation Metagenomics: Methodological Challenges and Opportunities. While Shotgun metagenomic sequencing requires 1 ng DNA input in minimum, 16S/ITS sequencing is much more sensitive with input minima being femtograms or even as low as 10 copies of 16S rRNA genes. Please contact us before purchasing your own supplies for qPCR to ensure you are using the correct type. Novogene offers comprehensive metagenomics solutions, from the study of community structure and species classification to system evolution, gene function, and metabolic network of environmental microorganisms. size ~550 bp, best ~450-500 bp for MiSeq; no size limit for Sequel 2) can be adapted to our systems. The library preparation workflow is similar to regular whole genome sequencing, including random fragmentation and adapter ligation. Biphenyl-utilizing bacteria isolated from the rhizosphere of the three different plant species showed low phylogenetic diversity and well represented functional traits, in terms of PGP and bioremediation potential. Extra costs may be incurred if samples, due to their nature, require more specialized (length, cost, etc.) 2 Yield estimates reflect optimum values defined by Illumina and apply to GSL prepared libraries only. We will try and point you to a suitable, Current GSL Services & Pricing (effective April 15th, 2022), 10X Genomics 3 Single Cell Library Prep services, Platinum Sequencing - PCR clean up (ExoSap), Seq Rxn, Clean Up & Capillary Run, Gold Sequencing - Seq Rxn, Clean Up & Capillary Run, Bronze Sequencing - Clean Up & Capillary Run, 10X Genomics 3' SC RNA library prep (internal users only, BSL1 only), NextSeq 500 v2 150 bp SR/ 75 bp PE flow cell, Per sample Sequencing (NovaSeq S4 150 PE), NovaSeq 6000 SP 100 bp SR/50 bp PE flow cell, NovaSeq 6000 S1 100 bp SR/50 PE flow cell, NovaSeq 6000 S2 100 bp SR/50 PE flow cell, $275.00 (caveats apply, contact GSL for more information), TapeStation DNA Analysis (D1000/D5000 Tapes), TapeStation High Sensitivity DNA Analysis, TapeStation High Molecular Weight DNA Analysis, BioAnalyzer RNA Analysis - Pico sensitivity, BioAnalyzer RNA Analysis - Nano sensitivity. sequencing costs per sample, but MiSeqs (or equivalent) are widely used for Metagenomic species profiling using universal phylogenetic marker genes. May the microbes be ever in your favor. Large sequencing least 6 wells of each 96-well plate for running positive/negative controls. This is why 16s rRNA sequencing is often referred to as targeted resequencing, this method targets the 16s rRNA gene only (For more information on 16s rRNA sequencing). projects that would have once cost as much as an apartment in San Francisco can contact us for custom pricing on any project. We can help people do this type of work but dont do such things as a service per se (interested in collaborations where we are involved in the science). Vergani, L., Mapelli, F., Marasco, R., Crotti, E., Fusi, M., Di Guardo, A., Borin, S. (2017). Any batches >380/190 will need to be spread across multiple MiSeq/Sequel 2 runs. Zuniga-Montanez R, Coil DA, Eisen JA, Pechacek R, Guerrero RG, Kim M, Shapiro K, Bischel HN. This is why it is recommended to use shotgun metagenomic sequencing for human-microbiome-related samples, such as feces and saliva, if taxonomy profiling is the main purpose. metagenomics sequencing throughput Why 90 samples? microbial communities. ): So we havent even gotten to the bioinformatics and other 3. Metagenomic species profiling using universal phylogenetic marker genes. metagenomic metadata antibiotic frameworks Metagenomic studies can be completed with a variety of metagenome sequencing methods including: Is there a difference between 16s rRNA sequencing and Shotgun metagenome sequencing? substantial amount of time (and mental anguish) to read poor handwriting on 10k CCS=HiFi reads/sample, MiSeq Standard Run (~20-25 M PE reads & 12-15 Gb), MiSeq Micro 2x150 bp Run (~4 M PE reads & 1.2 Gb), MiSeq Nano 2x150 bp Run (~1 M PE reads & 0.3 Gb), NextSeq Sequencing Run (~400 M PE reads = 800 M single reads & 120 Gb), Library Prep. --Long read sequencing up to 8,000bp (or more) available with the PacBio Sequel! For example, the ZymoBIOMICS Spike-in Control I contains two microbes alien to the human microbiome (Imtechella halotolerans and Allobacillus halotolerans), whose genomes were previously not available. people doing the lab work already know exactly what they are doing1: PCR amplification (duplicate 25 L reactions): Gel electrophoresis (you want to make sure the PCRs worked, Kim D, Song L, Breitwieser FP, Salzberg SL. instrument can generate 300 million reads per lane at a cost of ~$2000 per lane. small tubes and keep track of what samples are going into what wells. We are proud to be able to support your research by generating high-quality, publication-ready data in a rapid time-frame. Similarly, ITS sequencing follows the same strategy but targeting the ITS (Internal transcribed spacer) region found in fungal genomes. The taxonomy resolution of 16S/ITS sequencing depends on the variable regions targeted, the organism itself, and the sequence analysis algorithm. In contrast to whole sequencing and targeting all genes from only one organism, shotgun metagenome sequencing is able to target all genes from ALL organisms found in an environmental sample. Kraken2 and Centrifuge3) or selected marker genes (MetaPhlAn4 and mOTU5) to generate a taxonomy profile. Details of our amplicon and metagenomics pipelines are available at https://github.com/mlangill/microbiome_helper, but a summary of the major deliverables clients will receive (analyses will require a "mapping file" from the clients containing any relevant metadata for the study) are available on our Protocols page. prior to DNA extraction. liquid-handling robots. To use an example, lets walk through a hypothetical budget because we often do Reach out to us and we will get back to you shortly. Shotgun metagenome sequencing is able to provide scientists a more full picture of the microbial diversity present in their environmental samples. solve that problem. Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Note:All prices below are for all researchers, including those affiliated with NCSU, UNC, Duke, and other federally-funded or non-profit organizations. Thus, although the declining cost of sequencing is Not only does Shotgun metagenome sequencing provide the ability for researchers to gain insight into the taxonomic makeup of their environmental samples, but because this method is not limited to the 16s rRNA gene, researchers are also able to gain insight into the functional genes present as well as the metabolic pathways. This stems from the species coverage of available reference databases because the taxonomy prediction of these sequencing approaches heavily depends on the reference database used. DADA26, have dramatically improved the accuracy and taxonomy resolution of this technique. metagenomics Im also assuming that On the other hand, shotgun metagenome sequencing targets all genes from all microbial organisms. At my university, this is equivalent to a take home annual salary of $44K But, with shotgun metagenomic sequencing, unless there is a perfect representative genome in the reference database for a microbe sequenced, the bioinformatics analysis is likely to predict the existence of multiple closely-related genomes. The chemodiversity of paddy soil dissolved organic matter correlates with microbial community at continental scales. Bacteria Associated to Plants Naturally Selected in a Historical PCB Polluted Soil Show Potential to Sustain Natural Attenuation. Im using amplicon sequencing here because it is a sequencing dna megabase borrelia metagenomic rrna ixodes 16s ricinus ticks
I agree that Novogene Corporation may use this information to contact me to assist with my request. reasons that are too complicated to go into here) and we typically reserve at Last but not least it is worth reiterating that I havent the lab does not have to purchase any of the equipment required (e.g. metagenomic functional dna extracted reservoirs uncovered investigations Centrifuge: rapid and sensitive classification of metagenomic sequences. 16S sequencing or shotgun sequencing? We would like to sequence the whole metagenome of different microbial consortia, can you provide the service to external institutions? This is demonstrated when sequencing DNA from the mock microbial community (e.g. metagenomics microbial sequencing steps science revealing secrets planet project nap edu Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. On the other hand, if analyzed with 16S sequencing, they will be identified due to the presence of their 16S sequence in reference databases. Sunagawa S, et al. The Beauty of Science is to Make Things Simple, How to get quality DNA for ChIP Sequencing, How to Sequence RRBS Libraries on Illumina, Learn how to sequence RRBS libraries on Illumina platforms, How to Extract RNA from Hard-to-Lyse Samples in Trizol, Learn the best extraction methods for challenging sample types. PGP traits, including 1-aminocyclopropane-1-carboxylic acid deaminase activity potentially associated to plant stress tolerance induction, were widely distributed among the isolates according to in vitro assays. But on way to office from there saw a spectacular American Kestrel and wanted to share. These are valid questions and questions that can be resolved with a quick breakdown of metagenome sequencing methods. metagenomic pipeline datasets metagenomics microbial + Sequencing (~50 Gb/genome), Library Prep.
depending on what is required) and we are already at >$1600 for the 90 Change), You are commenting using your Facebook account. LAB HOME SERVICES INSTRUMENTS SUBMISSIONS Contact Us. I understand that all personal information I have submitted will be kept confidential in accordance with Novogene's privacy policy. Wet lab to justify far higher data output per run. The GSL is a University BSL-1 laboratory, classified as a teaching and training facility with student interactions. no false positives. In order to ensure the accuracy and reliability of the sequencing data, quality control (QC), consistent with our high scientific standards, is performed at each step of the procedure. Because of this, 16S/ITS sequencing is better in regard to false positives. The cost estimates are often a bit surprising + Sequencing (~50 Gb/sample), Library Prep. disseminated plot illustrating the declining cost of sequencing has to be In short you can If composition profiling is the main purpose of the study, both techniques have pros and cons to be considered (Table 1). metagenomic microbial metagenomics concepts These closely related genomes can be from different species of the same genus or even different genus. been collected and are sitting in your lab freezer organized and ready to go. Consequently, the former has greater cross-domain coverage. extraction procedures. https://github.com/mlangill/microbiome_helper, Library Preparation + Sequencing (max. Gut Microbes, 1-13. Li, H. Y., Wang, H., Wang, H. T., Xin, P. Y., Xu, X. H., Ma, Y., & Arnold, W. (2018).
Kim D, Song L, Breitwieser FP, Salzberg SL. This one was seen recently in Davis, CA, Oystercatcher doing its thing / in Berkeley CA, Questions for Jonathan Eisen (Advice, Background, Projects, etc), SWAN (Studies of Womens Health Across the Nation) Microbiome, Genomic Encyclopedia of Bacteria and Archaea (GEBA), Publications: Animal-Microbe Interactions, Publications: Phylogenomic and Computational Methods and Tools, Publications Marine Symbioses and Microbiology, Publications Phylogenetic Diversity of Genomes, Publications: Plants and Associated Microbes, Jonathan Eisens TED Talk on Meet Your Microbes, Data from Morgan et al. Just got out of a meeting with @ucdavis Chancellor @chancellor.may and his leadership group. On the other hand, the interference of host DNA is a much more difficult problem for shotgun metagenomic sequencing even though the cost of sequencing has decreased dramatically. Im assuming here that everything works well the + Sequencing (~4 M PE reads = 8 M single reads & 1.2 Gb/sample), Library Prep. doi: 10.1371/journal.pone.0267212. One control well is required per 96-well PCR library plate, hence a maximum of 4x95=380 samples can be done together on one MiSeq run (190 for a 2X run) or 2x95=190 samples on a Sequel 2 run (95 for a 2X run). Once you have our work in 96-well plates(I dont trust 384-well pipelines for Effects of spaceflight on the composition and function of the human gut microbiota, Gut MicrobesIssue Date: 2020 Jul 3IF: 7.740DOI: 10.1080/19490976.2019.1710091. Centrifuge: rapid and sensitive classification of metagenomic sequences. Nature Biotechnology 2013 31(9): 814-821.
+ Sequencing (~0.25 M PE reads = 0.5 M single reads & 150 Mb/genome), Library Prep. sequencing itself is often a small fraction of the total cost of the microbial Send email to jonathan dot eisen at gmail dot com. 4. costs), $115 in reagents/consumables (SequalPrep plates + Sequencing (~12 Gb and ~500k reads/sample). These sample So, Right? DNA sequencing is clearly a powerful tool for analyzing All the rhizosphere communities, nevertheless, hosted bacteria with degradation/detoxification and PGP potential, putatively sustaining the natural attenuation process. Currently, the coverage of 16S/ITS databases is much better than whole-genome databases. Users are charged in increments of 0.25 hours, depending on total use time. On average, 72% of the strains harbored the bphA gene and/or displayed catechol 2,3-dioxygenase activity, involved in aromatic ring cleavage. The presence of too much host DNA can cause non-specific amplification in the library preparation process of 16S and ITS sequencing, but the impact is controllable by adjusting PCR cycles and changing primers. After all, an Illumina HiSeq 4000 4 Block sequencing is performed on NovaSeq S4 150 PE lanes, and is only available for select GSL-prepared library options, we cannot offer block pricing options for user prepared libraries, or those not suitable for sequencing at 1502 PE .
of the total cost. Dr. Rojas got a UC Davis Chancellors Postdoctoral Fellow Award to come to UC Davis to work on cats and the role of Continue reading "Article about Dr. Conni []. Unlike a fictional lab document.getElementById( "ak_js_1" ).setAttribute( "value", ( new Date() ).getTime() ); Enter your email address to follow this blog and receive notifications of new posts by email. This exciting development has made it possible to achieve complete maps of certain bacteria and fungi for the purposes of taxonomy resolution and functional annotation. 3: Per-sample costs can go up appreciably if one 4: Liquid-handling robots are expensive, they - 2x150bp 10-20 million paired sequences per sample = $400 (data only) Illumina NovaSeq, -2x300bp 1-2 million sequences = $600 (data only) Illumina MiSeq, -2x250bp 10 million sequences = $650(data only) Illumina HiSeq, -2x250bp 4 million sequences = $450 (data only) Illumina HiSeq. The cost of making 24 (meta)genomic libraries, using the TruSeq kit (preferred for high-input samples) and pooling them for a single MiSeq PE250 run is approximately $2600. Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. If you wish to have more data or less data per sample feel free to contact us and we can see how prices are affected. bit easier to estimate the costs and because such methods are used by many labs A typical workflow for taxonomy analysis of shotgun metagenomic data includes quality trimming and comparison to a reference database comprising whole genomes (e.g. Here presents a list of academic publications that have used Novogene Shotgun Metagenomic Sequencing services. After sequencing, raw data is analyzed with a bioinformatics pipeline which includes trimming, error correction, and comparison to a 16S reference database. water, or bellybutton lint and use DNA sequencing-based approaches to analyze 2 When running the CFX384 qPCR machine, the user must use BioRad optical seals and qPCR plates. What is the difference between Metagenomics and Metagenome Sequencing? To learn more please see our Privacy Policy. PMID: 35452479; PMC [], New preprint out on from the Eisen Lab and others.
metagenomics enables Whats the take home message here? 2022 Zymo Research Corporation. This is likely nave4. + Sequencing (~2 Gb/genome), Library Prep. 50k reads/sample, Standard Bioinformatics Analysis Pipeline, Library Preparation + Sequencing (max. 3 Lane or block pricing reflects divided user costs, however sequencing turn-around times are subject to sufficient user volume in order to fill the flow cell. 2: These cost estimates are for a MiSeq run. This sequencing method is aptly named "Shotgun" due to its widespread of targets, whereas targeted resequencing methods could by the same logic be named "Rifle" metagenome sequencing.